Histone Recycling Maintains the Epigenome
Published in Molecular Cell, a study from the Groth laboratory at the Biotech Research and Innovation Centre, University of Copenhagen, shows that during DNA replication histone modifications, a crucial component of the epigenome, are transferred from unreplicated DNA to the same genomic location on replicated DNA with surprisingly high accuracy, but that after replication, the speed with which original modification levels are restored is not uniform throughout the genome.
Though the sequence of the genome is the same in almost all cells in our body, cell types differ widely. The morphology and function of a cell is determined by which genes from the genome are expressed and which are not. Gene expression is regulated by the epigenome, proteins and chemical marks that package and decorate the genome in unique patterns between different cell types. For cell identity to be maintained between cell divisions, this unique pattern must be reproduced on new DNA, but how this occurs remains unclear.
To further understand how the epigenome is copied onto new DNA during DNA replication, researchers in the Groth lab focused their study on histone modifications, one of the best-studied epigenetic marks. To specifically profile the location of histone modifications on newly replicated DNA, they developed a new technology, Chromatin Occupancy after Replication (ChOR-seq). Using ChOR-seq, they compared the position of different histone modifications before and after DNA replication.
The epigenome withstands the disruption caused by DNA replication
Regardless of the kind of histone modification profiled, it was found in approximately the same position before and after DNA replication. The researchers designed their study to specifically track deposition of parental histones, the histones that had previously packaged the unreplicated genome, by profiling histone modifications only found on these. This result therefore shows that modified parental histones, evicted from DNA during DNA replication, are “recycled”, or re-incorporated, at or very close to their original position. Professor Anja Groth says, “It is really exciting that we now show that positional information in histone modifications is inherited during DNA replication. This is a prerequisite for these modifications serving an epigenetic function, and thus an important step towards resolving how histone-based memory contributes to cell fate decisions.”
The researchers next inhibited EZH2, a “writer” enzyme that establishes a modification called H3K27 trimethylation on histones, and monitored H3K27 trimethylation throughout the cell cycle. They found that genomic regions typically containing H3K27 trimethylation remained stably marked with the modification up to 24 hours post-replication. Therefore, for H3K27 trimethylation, recycled histones alone were sufficient to create stable domains of the genome decorated with this modification. The epigenome is therefore unexpectedly resilient against the disruption DNA replication imposes on it.
The speed of full epigenome re-establishment is modification- and locus-specific
Because there is the double amount of DNA after replication, the recycling of modified histones is complemented with deposition of new naïve ones, and the overall level of modifications decreases by half on replicated DNA relative to unreplicated DNA. Researchers in the Groth laboratory therefore next used their ChOR-seq technology to ask if the re-establishment of histone modification levels is uniform throughout the genome. This was not the case- certain regions of the genome had their modification levels restored faster than others. These fast-restoring regions were those that best attract the “writer” enzymes that re-establish histone modifications. Further, modifications were restored with different speeds for different marks - some returned to unreplicated levels before cell division, and some returned to unreplicated levels only in daughter cells.
One of the lead authors of the study, Nazaret Reverón-Gómez, says, “The epigenome is generally thought of as static within a population of cells of the same type. This study disproves that idea, and instead shows that DNA replication and progression through the cell cycle create oscillations in modification levels genome-wide. The epigenome therefore varies considerably between cells, even cells of the same type, based on their cell cycle stage.” These variations have the potential to influence cell identity, both in a regulated manner, as in development, and in disease states such as cancer. In the future, it will therefore be important to determine the contribution of cell cycle-induced epigenomic variations in these biological processes.
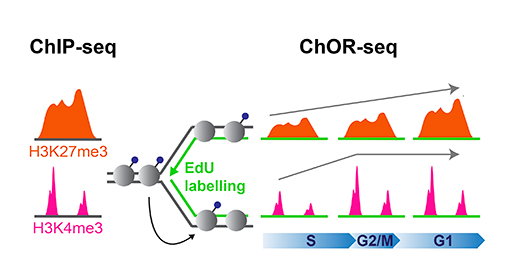
Prior to replication, regions in the genome are marked with histone modifications, such as H3K27me3 and H3K4me3. Post-replication, positional information is maintained through accurate histone recycling but the modifications are diluted relative to DNA and restored to pre-replication levels over time with distinct kinetics.
Contact
Professor Anja Groth
Email: anja.groth@bric.ku.dk
Phone: +45 35 32 55 38
Communications officer Anne Rahbek-Damm
Email: anne.rahbek@bric.ku.dk
Mobile: 21 28 85 41
Publication
Reverón-Gómez N, González-Aguilera C, Stewart Morgan KR, Jakobsen JS, Alabert C, Groth A (2018) Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication.https://www.sciencedirect.com/science/article/pii/S1097276518306440?via%3Dihub
