Unique oncogenetic programmes influence response to drug treatment in bile duct cancer patients
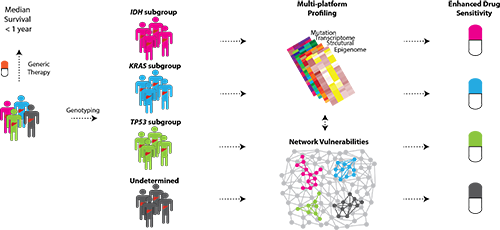
In a large study from the University of Copenhagen, researchers have shown that a single-gene dissection approach can discriminate divergent cancer programs and drug vulnerabilities in patients with intrahepatic cholangiocarcinoma. Their stratification of patients based on the mutational status of three individual classifier genes (IDH1/2, KRAS, TP53) uncovered unique oncogenic programs at play that influence response to drug treatment. The results have just been published in Hepatology.
This study is novel as it probes the genomic, epigenomic and pharmacologic landscapes of a disease that is characterized by extensive molecular heterogeneity and innate chemoresistance. While the immediate roles of these classifier genes are well established in oncology, the network-wide implications of having a mutation in any of these genes, as uncovered in this study, are completely unprecedented. Jesper B Andersen, research leader
The researchers from the Andersen Group at Biotech Research & Innovation Centre (BRIC) analyzed genome-wide data obtained from 496 intrahepatic cholangiocarcinoma (iCCA) patients. From these analyses, the team elucidated unique mutational signatures, co-mutational spectra, deregulated signaling pathways, structural alterations and DNA methylation aberrations associated with each patient subgroup. To test the clinical implications of such unique oncogenic programs, they utilized a drug repositioning approach and screened a library of 525 drugs in patient-matched cell models.
These findings uncover the potential of individual mutations to induce substantial downstream molecular heterogeneity which could facilitate prediction of therapeutic sensitivities for iCCA patients using standard targeted genotyping.
Significant implications for utility of precision medicine
Given the clinical and biological heterogeneity of iCCA, it has become abundantly clear that precision medicine will be the most effective method to reduce growing mortality rates for this disease. In spite of the gross differences in oncogenic programs observed in our study, the majority of patients are currently treated alike. By stratifying patients based on `omics´ using our classifier genes - with genotyping technology that is routinely used in the clinic -, an unprecedented level of insight into a patient’s cancer may be obtained with significant implications for development or utility of precision medicine, Jesper B Andersen
All 525 drugs tested in this study are either FDA-approved or at late-stage clinical development, and were tested in the patient-matched models at clinically-achievable doses. This indicates that the compounds are known to be safe in humans and are already available in clinics for patients with other malignant and non-malignant conditions.
It is a complex, time consuming and costly process to develop new therapies. Here, we take advantage of drugs already in clinical use and try to repurpose them for successfully treating iCCA patients, thereby streamlining the prospective clinical benefit of our study. Dr. Colm J. O’Rourke, first-author together with Dr. Chirag Nepal and postdoc in the Andersen group.
Next: in vivo testing
While patient-specific vulnerabilities in part recapitulated subgroup-associated pathway alterations predicted in silico, culture systems have well-noted shortcomings (no microenvironment or immune system, different physiological conditions) which may modify drug response. As such, the next stage will be to test these compounds in vivo using carefully controlled genetic models and patient transplants. Pending continued therapeutic validation in a genotype-guided clinical trials in collaboration with Danish and international clinical collaborators.
Info graphics. Click for large image
Contact
Jesper B Andersen, Group leader
Mail: jesper.andersen@bric.ku.dk
Cell phone: 0045 2151 6245
Anne Rahbek-Damm, communications officer
Mail: anne.rahbek@bric.ku.dk
Cell phone: 0045 2128 8541
Contact
Jesper B Andersen, Group leader
Mail: jesper.andersen@bric.ku.dk
Cell phone: 0045 2151 6245
Anne Rahbek-Damm, communications officer
Mail: anne.rahbek@bric.ku.dk
Cell phone: 0045 2128 8541
The paper
Genomic Perturbations Reveal Distinct Regulatory Networks in Intrahepatic Cholangiocarcinoma.
Nepal, O'Rourke CJ, Oliveira DV, Taranta A, Shema S, Gautam P, Calderaro J, Barbour A, Raggi C, Wennerberg K, Wang XW, Lautem A, Roberts LR, Andersen JB.
Hepatology. 2017 Dec 26. doi: 10.1002/hep.29764. [Epub ahead of print]
About iCCA
Intrahepatic cholangiocarcinoma is a rare malignancy originating in the bile ducts in the liver and is unfortunately among one of the few malignancies with increasing incidence and mortality rates worldwide. Tumors in the biliary system, such as iCCA, are deadly and currently with no approved or efficacious therapy besides surgical resection, a procedure that is only offered to 10-30% of patient’s due to typical locally advanced and metastatic disease presentation at diagnosis. Therefore, iCCA requires more emphasis to elucidate novel treatment strategies.