Bioinformatics
The BRIC Bioinformatics Core Facility supports researchers at BRIC, the University of Copenhagen (UCPH) and external institutions.
The Facility is composed of a multidisciplinary team updated on the latest technologies in molecular biology.
- consultation
- experimental desing
- data analysis, data management
- guidance
- short & long-term projects
- training.
Although you are welcome to contact us for other bioinformatic needs.
Consultation
You are always welcome to talk to us! We can work together to understand your needs.
No fee is applied to understand your needs
Experimental Desing
Including a bioinformatician in an early stage of your project can save you time, work, and money.
Data Analysis
We adapt to your needs. We can discuss and find together the best way to analyze your data. For more details on a typical bioinformatics project, please check section "Example of a standard bioinformatics projects" below.
Also, if you want to analyze the data yourself we can provide support for your bioinformatic analysis with your own code or with existing pipelines.
End-To-End
We aim to be able to offer an End-to-End service for your project. We work in close collaboration with other BRIC core facilities to offer and End-To-End service
Library Preparation - Sequencing - Bioinformatics
Please, Talk to us for more information on how to get the best of us.
More services
You can also request our services for Data Management. We can prepare and submit your data to any public database.
Don’t hesitate to contact us to help you preparing your manuscript or grant application.
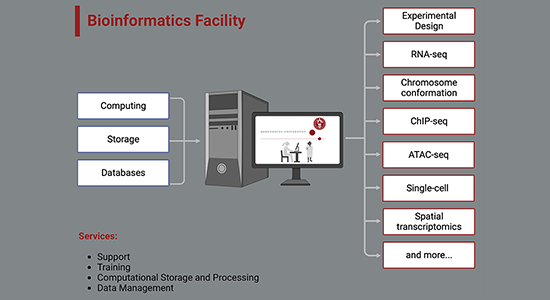
Access to a computing and storage infrastructure with
- Automated backup
- Software tools for formatting, analyzing, visualizing, interpreting and comparing biological data.
- Databases and knowledge bases storing biological knowledge.
Training programs on demand
On handling and analyzing data for users at BRIC with diverse backgrounds and experience.
A standard bioinformatic project would be a project with typically around 6 samples, 3 samples of each condition, where the user aims to perform a differential expression analysis between the two conditions and a functional enrichment analysis. The data types included in such category are:
- RNA-seq
- ChIP-seq
- ATAC-seq
- Single Cell data
Estimating hours is always challenging; We estimate a workload of 20 to 30 hrs for a standard analysis of simple datasets.
An Example of a Standard RNA-seq dataset analysis
- Initial meeting / communications
- Reading about the project
- Transfer raw sequencing data
- Quality assessment of the data
- Communication with the user
- Alignment of sequencing reads onto the genome of interest and gene quantification
- Differential gene expression analysis
- Functional enrichment
- Report and Data visualization
- Meetings to discuss results and possible follow-ups
A plus in using our services: We work with optimized pipelines ensuring reproducibility and providing reports.
BRIC users: 400 DKK/hour
UCPH users: 500 DKK/hour
External users: 980 DKK/hour
A Functional Link between Nuclear RNA Decay and Transcriptional Control Mediated by the Polycomb Repressive Complex. William Garland, Itys Comet*, Mengjun Wu*, Aliaksandra Radzisheuskaya*, Leonor Rib*, Kristoffer Vitting-Seerup, Marta Lloret-Llinares, Albin Sandelin, Kristian Helin, Torben Heick Jensen. Cell reports 2019 Nov 12; 29 (7), 1800-1811. e6
Rapid recapitulation of nonalcoholic steatohepatitis upon loss of host cell factor 1 function in mouse hepatocytes. Molecular and cellular biology. Minocha S, Villeneuve D, Praz V, Moret C, Lopes M, Pinatel D, Rib L, Guex N, Herr W. 2019 Mar 1;39(5):e00405-18.
Differential regulation of RNA polymerase III genes during liver regeneration. Yeganeh M, Praz V, Carmeli C, Villeneuve D, Rib L, Guex N, Herr W, Delorenzi M, Hernandez N, Deplancke B, Desvergne B. Nucleic acids research. 2018 Dec 29.
Cycles of gene expression and genome response during mammalian tissue regeneration. Rib L, Villeneuve D, Minocha S, Praz V, Hernandez N, Guex N, Herr W. Epigenetics & chromatin. 2018 Dec;11(1):52.
YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MR, Rundsten CF, Johansen JV, Li Y, Madsen CD, Nakamura T, Watanabe M, Nielsen OH, Schweiger PJ, Piccolo S, Jensen K. Cell Stem Cell. 2018 Jan 4;22(1):35-49.
Transcriptome profile of lung dendritic cells after in vitro porcine reproductive and respiratory syndrome virus (PRRSV) infection. Pröll MJ, Neuhoff C, Schellander K, Uddin MJ, Cinar MU, Sahadevan S, Qu X, Islam MA, Poirier M, Müller MA, Drosten C. PloS one. 2017 Nov 15;12(11):e0187735.
Segregated hepatocyte proliferation and metabolic states within the regenerating mouse liver. Minocha S, Villeneuve D, Rib L, Moret C, Guex N, Herr W. Hepatology communications. 2017 Nov;1(9):871-85.
CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1–Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Engelholm LH, Riaz A, Serra D, Dagnæs-Hansen F, Johansen JV, Santoni-Rugiu E, Hansen SH, Niola F, Frödin M. Gastroenterology 153 (6), 1662-1673. e10. doi: 10.1053/j.gastro.2017.09.008. PubMed PMID: 28923495.
Impaired removal of H3K4 methylation affects cell fate determination and gene transcription. Lussi YC, Mariani L, Friis C, Peltonen J, Myers TR, Krag C, Wong G, Salcini AE. Development. 2016 Oct 15;143(20):3751-62.
An interactive environment for agile analysis and visualization of ChIP-sequencing data. Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. Nat Struct Mol Biol. 2016 Apr;23(4):349-57. doi: 10.1038/nsmb.3180. PubMed PMID: 26926434.
Identification of gene co-expression clusters in liver tissues from multiple porcine populations with high and low backfat androstenone phenotype. Sahadevan S, Tholen E, Große-Brinkhaus C, Schellander K, Tesfaye D, Hofmann-Apitius M, Cinar MU, Gunawan A, Hölker M, Neuhoff C. BMC genetics. 2015 Dec;16(1):21.
H3K23me2 is a new heterochromatic mark in Caenorhabditis elegans. Vandamme J, Sidoli S, Mariani L, Friis C, Christensen J, Helin K, Jensen ON, Salcini AE. Nucleic acids research. 2015 Oct 17;43(20):9694-710.
Preliminary study of FMO1, FMO5, CYP21, ESR1, PLIN2 and SULT2A1 as candidate gene for compounds related to boar taint. Neuhoff C, Gunawan A, Farooq MO, Cinar MU, Große-Brinkhaus C, Sahadevan S, Frieden L, Tesfaye D, Tholen E, Looft C, Schellander K. Meat science. 2015 Oct 1;108:67-73.
Exploring novel mechanistic insights in Alzheimer’s disease by assessing reliability of protein interactions. Malhotra A, Younesi E, Sahadevan S, Zimmermann J, Hofmann-Apitius M. Scientific reports. 2015 Sep 8;5:13634.
MicroRNA expression profile in bovine granulosa cells of preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. Gebremedhn S, Salilew-Wondim D, Ahmad I, Sahadevan S, Hossain MM, Hoelker M, Rings F, Neuhoff C, Tholen E, Looft C, Schellander K. PLoS One. 2015 May 19;10(5):e0125912.
Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. Genes Dev. 2015 May 1;29(9):910-22. doi: 10.1101/gad.260174.115. PubMed PMID: 25886910; PubMed Central PMCID: PMC4421980.
26 The Cellular Plasticity of Fetal and Adult Intestinal Epithelium and Its Functional Evaluation by Grafting Assay. Yui S, Guiu J, Schweiger P, Rundsten CF, Jensen K. Gastroenterology. 2015 Apr 1;148(4):S-9.
Loss of PRDM11 promotes MYC-driven lymphomagenesis. Fog CK, Asmar F, Côme C, Jensen KT, Johansen JV, Kheir TB, Jacobsen L, Friis C, Louw A, Rosgaard L, Øbro NF. Blood. 2015 Feb 19;125(8):1272-81.
Single site suppressors of a fission yeast temperature-sensitive mutant in cdc48 identified by whole genome sequencing. Marinova IN, Engelbrecht J, Ewald A, Langholm LL, Holmberg C, Kragelund BB, Gordon C, Nielsen O, Hartmann-Petersen R. PloS one. 2015 Feb 6;10(2):e0117779.
Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Mol Cell. 2014 Aug 7;55(3):347-60. doi: 10.1016/j.molcel.2014.06.005. PubMed PMID: 24999238.
Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Wu X, Johansen JV, Helin K. Mol Cell. 2013 Mar 28;49(6):1134-46. doi: 10.1016/j.molcel.2013.01.016. PubMed PMID: 23395003.
A genetic screen identifies BRCA2 and PALB2 as key regulators of G2 checkpoint maintenance. Menzel T, Nähse-Kumpf V, Kousholt AN, Klein DK, Lund-Andersen C, Lees M, Johansen JV, Syljuåsen RG, Sørensen CS. EMBO Rep. 2011 Jul 1;12(7):705-12. doi: 10.1038/embor.2011.99. PubMed PMID: 21637299; PubMed Central PMCID: PMC3128973.
TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. Nature. 2011 May 19;473(7347):343-8. doi: 10.1038/nature10066. PubMed PMID: 21490601; PubMed Central PMCID: PMC3408592.
JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. Nature. 2010 Mar 11;464(7286):306-10. doi: 10.1038/nature08788. PubMed PMID: 20075857.
RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Doehn U, Hauge C, Frank SR, Jensen CJ, Duda K, Nielsen JV, Cohen MS, Johansen JV, Winther BR, Lund LR, Winther O, Taunton J, Hansen SH, Frödin M. Mol Cell. 2009 Aug 28;35(4):511-22. doi: 10.1016/j.molcel.2009.08.002. PubMed PMID: 19716794; PubMed Central PMCID: PMC3784321.

Leonor Rib, PhD
I’m a Computer Scientist with a passion for biology.
I hold a PhD in computational biology after doing a PhD in Integrated experimental and computational biology in Switzerland.
I have extensive experience in the experimental design and analysis of multiomics and genetic data for more than 15 years in collaboration with hospitals, academic research groups and biotech companies.
More details can be found in my LinkedIn profile and Google scholar.
I’m originally from Spain, in Catalonia. And if you offer me a “Calçotada” in good company and a good wine with “Porró” you will make me happy :)
Contact

Leonor Rib, PhD
leonor.rib@bric.ku.dk
BRIC - Biotech Research & Innovation Centre
Ole Maaløes vej 5
DK-2200 Copenhagen
Office: Biocenter 1-3-20
