Weischenfeldt Group
 We are interested in the mutational mechanisms and clonal evolution of cancer, in particular mechanisms of complex structural variants and the impact on 3D chromatin organization.
We are interested in the mutational mechanisms and clonal evolution of cancer, in particular mechanisms of complex structural variants and the impact on 3D chromatin organization.
Cancer is a disease of the genome, fueled by the accumulation of genomic alterations that improve the fitness of the cell.
We use computational cancer genomics approaches to study clonal evolution and cancer stem cell models, with the overarching goal to understand the mutational processes that allow tumor cells to escape and evolve in time and space to cause treatment-resistance.
We have a particular interest in the larger complex structural variants, their presence and impact on cancer and clonal evolution, and how they impact the chromatin architecture.
Structural variant mediated chromatin alterations and enhancer hijacking mechanisms in cancer genomes
- Dubois, Sidiropoulos et al, Nature Reviews Genetics, 2022
- Sidiropoulos et al, Genome Research, 2022
- Rheinbay et al, Nature, 2020
- Weischenfeldt et al, Nature Genetics, 2017
- Northcott et al, Nature, 2017
Evolutionary and mechanistic mutational forces shaping cancer genomes
- Li et al, Nature, 2020
- PCAWG consortium, Nature, 2020
- Hansen et al, Blood, 2019
- Gerhäuser et al, Cancer Cell, 2018
- Gröbner et al, Nature ,2018
- Rausch et al, Cell, 2012
Age-dependent mutational processes in cancer genomes
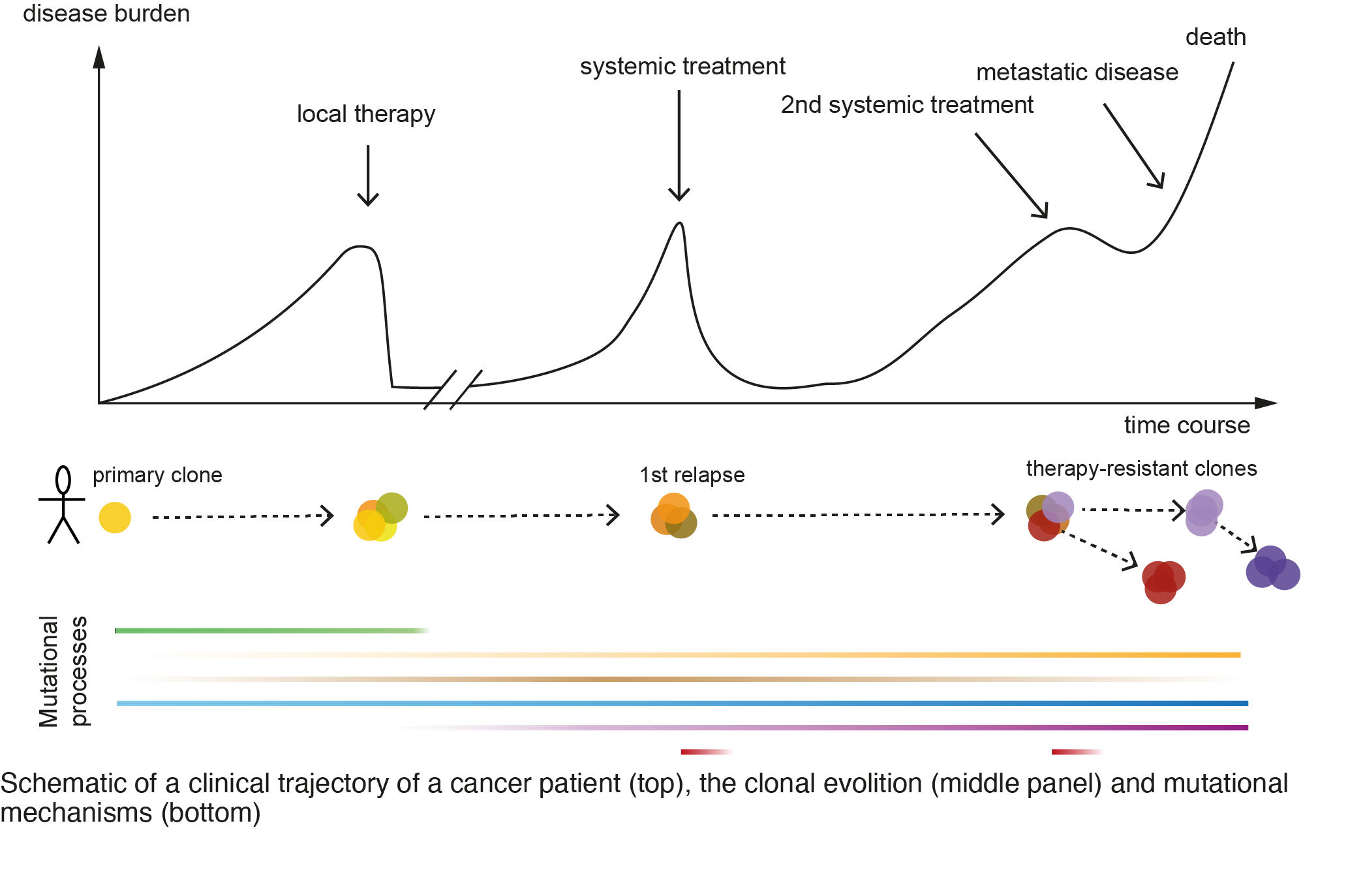
Clonal evolution analysis to identify recurrent mechanisms of treatment resistance and identify novel therapeutic targets
- Predict clinical trajectories from cancer patients by developing methods to i) reconstruct and model the clonal evolution, ii) analyze clonal dynamics and interplay with tumor microenvironment, with the overarching goal to study mechanisms of cancer stem cells.
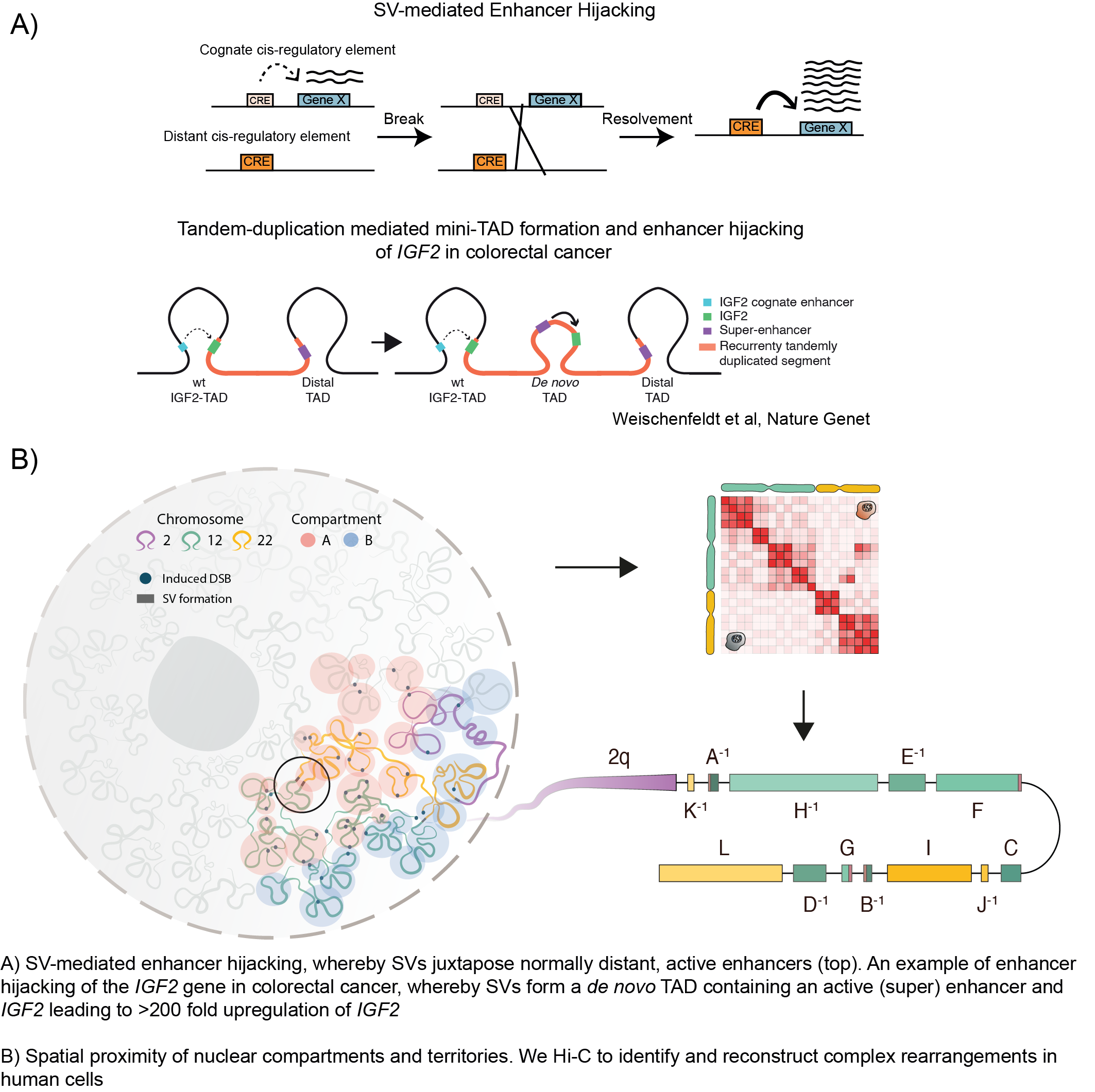
 How complex structural variants emerge and affect the 3D chromatin architecture in cancer
How complex structural variants emerge and affect the 3D chromatin architecture in cancer
- Integrate epigenetic with somatic alterations and patient-related data using data-driven quantitative genetics-based computational approaches.
- Apply chromatin architecture methodologies such as Hi-C to identify how genomic alterations can impact gene regulation through altered chromatin looping
We use a combination of computational and molecular biology technologies to reach our research aims. We have optimized and integrated a range of different sequencing approaches on clinical material including paired-end sequencing, RNA-seq, ATAC-seq, Hi-C, Capture-Hi-C, mate-pair seq, DNA panel seq, single-cell sequencing, long-read sequencing. We carry out our compute on HPC systems, primarily computerome.dk. We use git for code version control and Docker to deploy our code on other systems.
Selected publications
Structural variations in cancer and the 3D genome. # Dubois, F., Sidiropoulos, N., Weischenfeldt, J., and Beroukhim, R. 2022. Structural variations in cancer and the 3D genome. Nat Rev Cancer. DOI:10.1038/s41568-022-00488-9 (joint corresp author).
Somatic structural variant formation is guided by and influences genome architecture. # Sidiropoulos, N., Mardin, B. R., Rodríguez-González, F. G., Bochkov, I. D., Garg, S., Stütz, A. M., Korbel, J. O., Aiden, E. L., and Weischenfeldt, J. 2022. Genome Res 32: 643–655. DOI:10.1101/gr.275790.121 (first author, joint corresp author).
Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. # Rheinbay, E., Nielsen, M. M., Abascal, F., Wala, J. A., Shapira, O. et al. Nature, 578(7793), 102-111. doi:10.1038/s41586-020-1965-x (joint corresp author).
Patterns of somatic structural variation in human cancer genomes. # Li, Y., Roberts, N. D., Wala, J. A., Shapira, O., et al. Nature, 578(7793), 112-121. doi:10.1038/s41586-019-1913-9 (joint corresp author).
Molecular Evolution of Early-Onset PCa Identifies Molecular Risk Markers and Clinical Trajectories. # Gerhauser, C., Favero, F., Risch, T., Simon, R., Feuerbach, L., Assenov, Y., et al. Cancer Cell, 34(6), 996-1011.e8. doi:10.1016/j.ccell.2018.10.016 (joint first, and corresp author).
Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. * Weischenfeldt, J., Dubash, T., Drainas, A. P. et al. Nat Genet, 49(1), 65-74. doi:10.1038/ng.3722 (joint first author).
Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset PCa. * Weischenfeldt, J., Simon, R., Feuerbach, L., Schlangen, K., Weichenhan, D., Minner, S., et al. Cancer Cell, 23(2), 159-170. doi:10.1016/j.ccr.2013.01.002 (joint first author).
In the media
Studying DNA rearrangement to understand cancer
05 February 2020 - EurekAlert.org
Scientists identify new genetic drivers of cancer
05 February 2020 - EurekAlert.org
Global study maps cancer mutations in large catalogue
05 February 2020 - University of Copenhagen
Stort kræft-forskningsprojekt kortlægger uudforsket del af vores DNA
05 February 2020 - Videnskab.dk
Danskere afslører mekanisme bag aggressive kræftformer
22 November 2016 - Videnskab.dk.
Article (in Danish) on how 'big data' can be used to understand how cancer genes are activated.
View Weischenfeldt group on Twitter
Collaborative Research Programs
- Pan-prostate cancer group https://panprostate.org
- DCCC Brain Tumor Center https://dcccbraintumor.dk
- DCCC Blod cancer https://www.dccc.dk/nationale-forskningscentre/dansk-forskningscenter-for-pracisionsmedicin-i-blodkraft/
- Hauptstadt Urologie https://hauptstadturologie.dk






The laboratory has been awarded the LEAF - Laboratory Efficiency Assessment Framework Silver certificate.









